This module creates energy level diagrams common to atomic physics as matplotlib graphics. The level structure is defined using networkx graphs.
Quick Usage
This package takes networkx directional graphs, which can be used to effectively define a system hamiltonian, and creates an energy diagram representing the system. The nodes of the graph represent the energy levels. The edges of the graph represent the couplings between levels.
Passing a simple graph to the basic level diagram constructor will produce a passable output for simple visualization purposes.
nodes = (0,1,2)
edges = ((0,1),(1,2))
graph = nx.DiGraph()
graph.add_nodes_from(nodes)
graph.add_edges_from(edges)
d = ld.LD(graph)
d.draw()
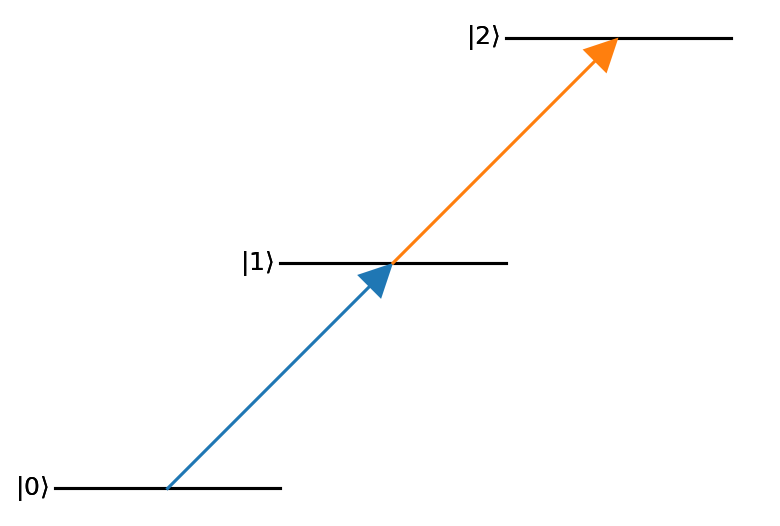
Global settings for the three primitive objects used by leveldiagram can be set by passing keyword argument dictionaries to the LD constructor.
To control options for a single level or coupling,
save these keyword arguments to the respective node or edge of the supplied graph.
Generally, the levels and couplings take standard matplotlib 2D line configuration arguments.
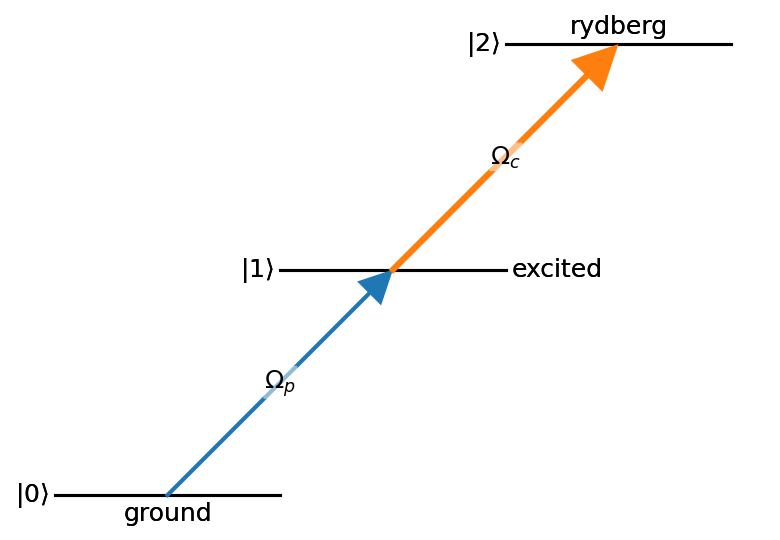
nodes = ((0,{'bottom_text':'ground'}),
(1,{'right_text':'excited'}),
(2,{'top_text':'rydberg'}))
edges = ((0,1, {'label':'$\\Omega_p$'}),
(1,2, {'label':'$\\Omega_c$'}))
graph = nx.DiGraph()
graph.add_nodes_from(nodes)
graph.add_edges_from(edges)
d = ld.LD(graph, coupling_defaults = {'arrowsize':0.15,'lw':3})
d.draw()
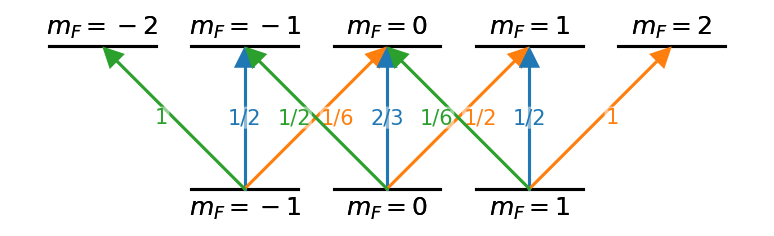
With some basic scripting to create the graph appropriately, much more complicated level diagrams can be made with relative ease.
hf_nodes = [((f,i), {('top' if f==2 else 'bottom') + '_text':'$m_F='+f'{i:d}'+'$',
'energy':f-1,
'xpos':i,
'width':0.75,
'text_kw':{'fontsize':'large'}})
for f in [1,2]
for i in range(-f,f+1)]
lin_couples = [((1,i),(2,i),{'label':l,'color':'C0',
'label_kw':{'fontsize':'medium','color':'C0'}})
for i,l in zip(range(-1,2), ['1/2','2/3','1/2'])]
sp_couples = [((1,i),(2,i+1),{'label':l,'color':'C1',
'label_offset':'right',
'label_kw':{'fontsize':'medium','color':'C1'}})
for i,l in zip(range(-1,2), ['1/6','1/2','1'])]
sm_couples = [((1,i),(2,i-1),{'label':l, 'color':'C2',
'label_offset':'left',
'label_kw':{'fontsize':'medium','color':'C2'}})
for i,l in zip(range(-1,2), ['1','1/2','1/6'])]
hf_edges = lin_couples + sp_couples + sm_couples
hf_graph = nx.DiGraph()
hf_graph.add_nodes_from(hf_nodes)
hf_graph.add_edges_from(hf_edges)
d = ld.LD(hf_graph, default_label = 'none')
d.ax.margins(y=0.2)
d.draw()